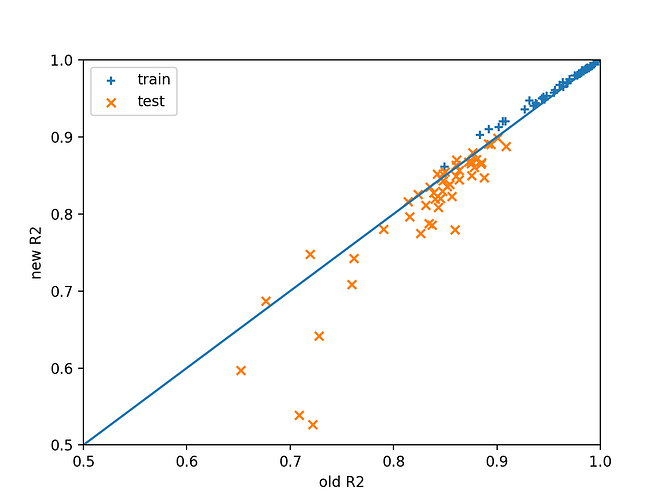
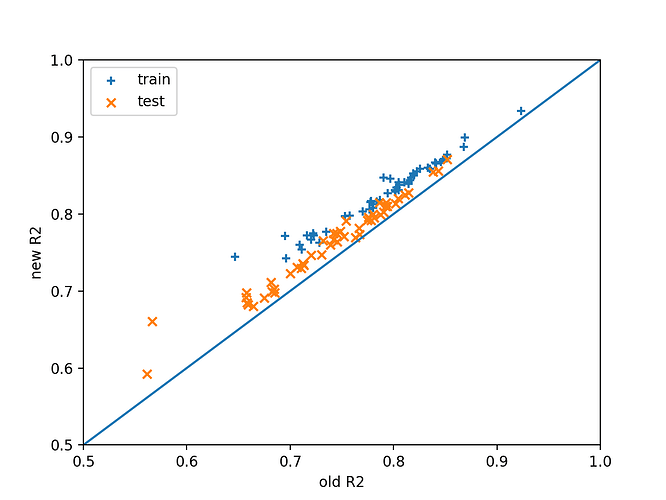
I did some investigations with regards to design matrix scaling. The question is whether we actually need this change to be implemented because it does break a lot of existing tests in the code. Note that two changes need to be made actually, applying to the case of “monte–carlo”, “user-defined” and “induced” meshes:
- Setting all quadrature weights to 1
- Post-multiplying by square root of “normalising factors”
= 1.0/np.sum(self.P ** 2, axis=1)
A quick test was set up to examine the difference caused by this change on least squares regression of an example function $f(x) = \sin (\sum_i x_i)$ with various input dimensions and maximum degrees of polynomial using user-defined meshes sampled from the uniform distribution between -1 and 1 in each dimension. The number of data points is set as twice the number of basis functions.
from equadratures import *
import numpy as np
from scipy.special import comb
import matplotlib.pyplot as plt
def f(x):
u = np.sum(x)
return np.sin(u)
d_range = range(3, 15)
p_range = range(2, 5)
seed_range = range(5)
all_r2 = np.zeros((len(seed_range), len(p_range), len(d_range)))
for t_ind, state in enumerate(seed_range):
for p_ind, p in enumerate(p_range):
for d_ind, d in enumerate(d_range):
# param and basis
my_params = [Parameter(distribution='uniform', lower=-1, upper=1, order=p) for _ in range(d)]
my_basis = Basis('total-order')
card = comb(p+d, d)
N = int(2.0 * card)
noise_var = 0.01
x = np.random.RandomState(state).uniform(-1, 1, (N, d))
y = np.apply_along_axis(f, 1, x) + np.random.RandomState(state).normal(0, noise_var, size=N).T
poly = Poly(parameters=my_params, basis=my_basis, method='least-squares',
sampling_args={'mesh': 'user-defined', 'sample-points': x,
'sample-outputs': y.reshape(-1, 1)})
poly.set_model()
N_test = 1000
x_test = np.random.RandomState(state).uniform(-1, 1, (N_test, d))
y_test = np.apply_along_axis(f, 1, x_test)
_, r2 = poly.get_polyscore(X_test=x_test, y_test=y_test)
print(state, p, d, r2)
all_r2[t_ind, p_ind, d_ind] = r2
The following results are obtained (test $R^2$ vs number of dimensions for various polynomial degrees). The dots are the old scaling, and dashed line new scaling.
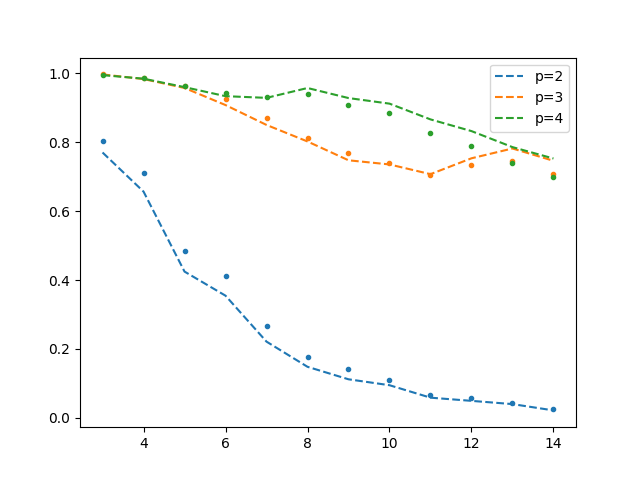
It can be seen that a non-negligible increase in $R^2$ is obtained using the new scaling for $p=4$. This agrees with the interpretation that this scaling is supposed to reduce instabilities with more pronounced effects when the design matrix is large (but perhaps not comprehensive enough to convince me that this is the case). I did not go further with the degree and dimension to test this because the running time of the script gets quite long. Perhaps this is more motivation to implement algorithms for solving large linear systems (e.g. iterative solvers)!