The ability to obtain predictive variance for Effective Quadratures’ polynomial approximations would be useful for a variety of applications. For example UQ of surrogate models, and guiding selection of suitable polynomial orders. Following section 1.4 in [1], for 1D training data with $N$ inputs $(x_1,\dots,x_N)$ and outputs $(y_1,\dots,y_N)$, a polynomial approximation can be obtained via least squares. For a polynomial of order $K$, the data can be vectorised
$$
\mathbf{X} =\left[\begin{array}{c} \boldsymbol{x}{1} \ \vdots\ \boldsymbol{x}{N} \end{array}\right] =
\left[\begin{array}{c} 1 & x_1 & x_1^2 & \dots & x_1^K \
\vdots & \dots & \vdots \
1 & x_N & x_N^2 & \dots & x_N^K \end{array}\right]
; ; ; ; \boldsymbol{y}=\left[\begin{array}{c}
y_{1}\ \vdots\ y_{N}
\end{array}\right],
$$
and, the polynomial approximation can be evaluated at new test points $\tilde{\boldsymbol{x}}$ with:
$$
\begin{align}
f(\tilde{\boldsymbol{x}};w)& \approx \tilde{\boldsymbol{x}}^T \left( \mathbf{X}^T\mathbf{X} \right)^{-1}\mathbf{X}^T\mathbf{y} \
&\approx \tilde{\boldsymbol{x}}^T\hat{\mathbf{w}} = \sum_{k=1}^K w_k x^k
\end{align}
$$
From section 2.10 in [1], the predictive variance at $\boldsymbol{\tilde{x}}$ is given by:
$$
\tilde{\sigma}^2(\tilde{\boldsymbol{x}}) = \widehat{\sigma^2} \tilde{\boldsymbol{x}}^T \left( \mathbf{X}^T\mathbf{X} \right)^{-1} \tilde{\boldsymbol{x}}
$$
Effective Quadratures does something similar, however with orthogonal polynomials:
$$
f(\tilde{\boldsymbol{x}};w)\approx \sum_{k=1}^K w_k p_k(\tilde{\boldsymbol{x}})
$$
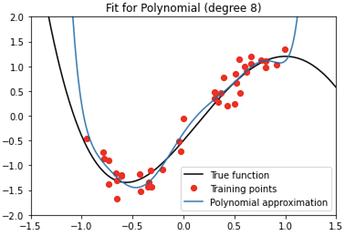
where Poly.get_coefficients() returns the vector polynomial coefficients $\boldsymbol{w}$, and Poly.get_poly() returns the polynomial basis functions $p_k(\tilde{\boldsymbol{x}})$. Poly.get_polyfit() evaluates the function $f(\tilde{\boldsymbol{x}};w)$, as shown in Figure 1.
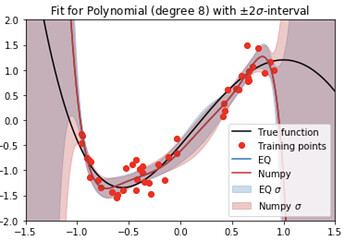
In this notebook, the aforementioned predictive variance calculation is extended to the EQ orthogonal polynomials. In Figure 2, the resulting $\pm 2\widetilde{\sigma}$ confidence intervals are added, and compared to the those obtained from numpy.polyfit().
Figure 2: 8th order 1D polynomial approximation from Effective Quadratures, with $2\tilde{\sigma}$ confidence intervals
Use cases
The obtained confidence intervals will be useful for determining uncertainty in our polynomial approximations. Additionally they may guide the selection of polynomial order, and in determining where additional training data is needed.
Todo’s
-
The predictive variance calculation is currently using the estimated (empirical) variance $\widehat{\sigma^2}$, which is equivalent to the root mean squared error at the training points (from Section 2.72 in [1]) $\widehat{\sigma^2} = \frac{1}{N}\sum_{n=1}^N \left(y_n - \boldsymbol{x}^T\hat{\mathbf{w}} \right)$. In some cases it might be preferable to specify the true variance $\sigma^2$, for example in cases where we know the mean and variance from experimental measurements, but don’t have discreet observations.
-
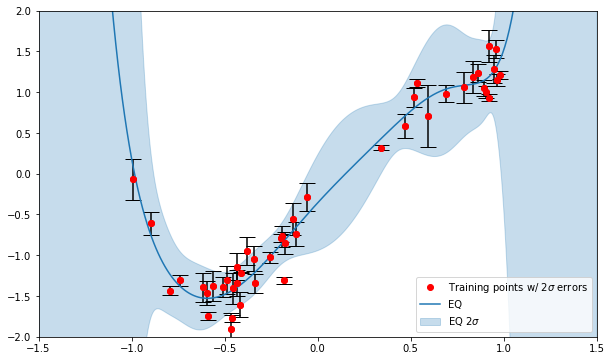
For 1D I have crudely validated by comparing to
numpy.polyfit(). I have implemented the above in 2D in this notebook but haven’t validated. Both could do with more rigorous validation (and unit tests). -
Merging into develop branch when ready.